While no treatments for the prevention of geographic atrophy (GA) occurrence or reversal of vision loss have so far proven fruitful, targeting lesion enlargement rate has seen some success with recently approved complement inhibitors such as pegcetacoplan and avacincaptad pegol. The genetics behind GA enlargement aren’t well understood, but they seem to “differ substantially” from GA susceptibility. Multiple conflicting reports have been published on the role of the ARMS2/HTRA1 genotype, which is strongly associated with increased GA risk. To reconcile these findings, researchers conducted a post-hoc analysis of an AREDS2 cohort to analyze this genotype as a risk factor for faster GA enlargement, based on change in area and progression to multifocality. The findings, published recently in Ophthalmology, revealed that GA area is a critical factor in the relationship.
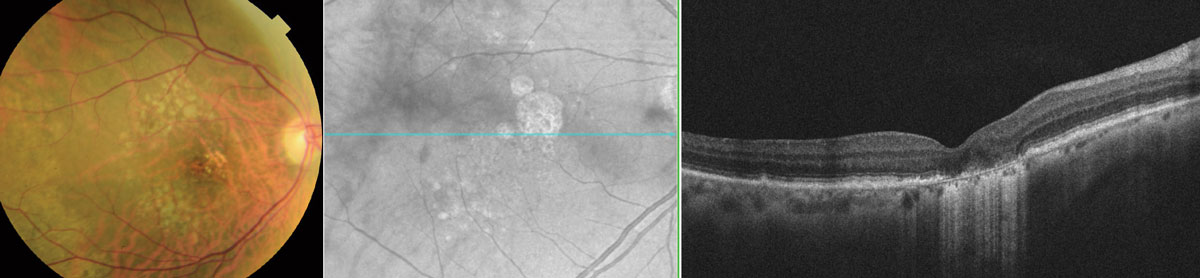 |
| Given that only small baseline GA areas were associated with faster enlargement, therapeutic strategies targeting ARMS2 or HTRA1 at the GA enlargement stage are unlikely to be successful for eyes with GA area above a certain size. Photo: Wendy Harrison, OD, PhD. Click image to enlarge. |
In the 406-participant cohort (546 eyes), researchers measured GA area from color fundus photographs taken at annual visits and performed statistical modeling to assess changes in square root GA area and progression to multifocality.
They found that GA enlargement was significantly faster in eyes with ARMS2 risk alleles, with rates of 0.224, 0.298 and 0.317 mm/year for 0 to 2 risk alleles, respectively. Additionally, they reported significant interaction between genotype and baseline GA area. Eyes with a small GA area (<1.9mm2) had significantly faster enlargement with ARMS2 risk alleles (0.193mm/year with 0 risk alleles vs. 0.304mm/year with 1 to 2 risk alleles). On the other hand, moderately small (1.9mm2 to 3.8mm2) or medium/large (≥3.8mm2) GA areas didn’t appear to be relevant risk factors for enlargement rate.
Eyes with non-multifocal GA had significantly faster enlargement with ARMS2 risk alleles, with rates of 0.175, 0.226 and 0.287mm/year with 0 to 2 risk alleles, respectively. The ARMS2 genotype wasn’t significantly associated with progression to multifocal GA.
The researchers concluded in their Ophthalmology paper that the ARMS2/HTRA1 genotype risk alleles represent a strong risk factor for faster GA enlargement only in eyes with very small GA area and a risk factor for “faster GA enlargement more through faster contiguous enlargement of existing lesions than through increased risk of additional new loci, towards multifocality.”
They explained in their paper that these risk alleles seem important in “increasing the risk of progression from non-advanced AMD to GA, and in initial GA enlargement (by contiguous enlargement), but not in subsequent enlargement, or in faster progression to multifocality.”
The researchers wrote that these findings might explain previously observed discrepancies in ARMS2/HTRA1 studies. “The results emphasize that the genetic risk factors for faster GA enlargement overlap very little with those for progression to GA. These findings have important implications for both routine practice and clinical trials, towards improved prognostic predictions of personalized enlargement rates and insights into the relevant biological pathways underlying GA enlargement.”
Agrón E, Domalpally A, Cukras CA, et al. Critical dependence on area in relationship between ARMS2/HTRA1 genotype and faster geographic atrophy enlargement: AREDS2 Report 33. Ophthalmology 2023. [Epub ahead of print]. |

